Brian Williamson, PhD
Biography
Brian Williamson, PhD, is a biostatistician with expertise in statistical epidemiology, semiparametric and nonparametric estimation theory, and high-dimensional estimation and prediction. He is interested in developing robust procedures for statistical inference when machine learning is used to address problems in public health, and in working toward accessible, affordable, high-quality healthcare for everyone. A central theme of his research is using prediction models (including machine learning and artificial intelligence) to make more accurate and efficient use of electronic health records data for research and clinical care.
Before joining Kaiser Permanente Washington Health Research Institute, Dr. Williamson completed his postdoctoral research training at the Fred Hutchinson Cancer Research Center. During his time at Fred Hutch, Dr. Williamson developed statistical methods to address issues arising in the development of biomarker panels for use in risk prediction, screening, and diagnosis. Dr. Williamson also collaborated with researchers from the Women’s Health Initiative to assess the utility of metabolomic biomarkers for predicting breast and colorectal cancer; with researchers from the HIV Vaccine Trials Network (HVTN) to aid in selecting candidate broadly neutralizing antibody regimens to advance to HIV prevention clinical trials; and was a part of the Coronavirus Prevention Network Biostatistics Team.
Dr. Williamson received his PhD in biostatistics from the University of Washington. His dissertation focused on a general framework for performing inference on model-free variable importance measures. With colleagues from the HVTN, he used this framework to identify features of the HIV viral genome that may be important in predicting viral susceptibility to the broadly neutralizing antibody VRC01.
At KPWHRI, Dr. Williamson collaborates on projects across a range of research areas including mental health, pragmatic clinical trials, and drug and vaccine safety and effectiveness.
Recent Publications
Minus E, Coley RY, Shortreed SM, Williamson BD Behavior of prediction performance metrics with rare events 2025 Nov 10;189:112046. doi: 10.1016/j.jclinepi.2025.112046. Epub 2025-11-10. PubMed
Williamson BD, King D, Huang Y Practical Considerations for Variable Screening in the Super Learner 2025 May 7 doi: 10.51387/25-nejsds82. Epub 2025-05-07. PubMed
Cohen P, Lambson BE, Mkhize NN, Moodley C, Yssel AEJ, Moyo-Gwete T, York T, Gwashu-Nyangiwe A, Ndabambi N, Thebus R, Juraska M, deCamp AC, Williamson BD, Magaret CA, Gilbert PB, Westfall D, Deng W, Mullins JI, Morris L, Williamson C, Moore PL Resistance mutations that distinguish HIV-1 envelopes with discordant VRC01 phenotypes from multi-lineage infections in the HVTN703/HPTN081 trial: implications for cross-resistance 2025 Feb 25;99(2):e0173024. doi: 10.1128/jvi.01730-24. Epub 2025-01-16. PubMed
Wolock CJ, Williamson BD, Shortreed SM, Simon GE, Coleman KJ, Yeargans R, Ahmedani BK, Daida Y, Lynch FL, Rossom RC, Ziebell RA, Cruz M, Wellman RD, Coley RY Importance of variables from different time frames for predicting self-harm using health system data 2024 Dec;160:104750. doi: 10.1016/j.jbi.2024.104750. Epub 2024-11-16. PubMed
Williamson BD, Huang Y Flexible variable selection in the presence of missing data 2024 Nov;20(2):347-359. doi: 10.1515/ijb-2023-0059. Epub 2024-02-13. PubMed
News
KPWHRI begins new phase of flu surveillance
KPWHRI receives $10 million to continue vaccine effectiveness research for flu, COVID-19, and other respiratory diseases.
Vaccine Safety

Biostatisticians track COVID-19 vaccine safety
Dr. Jennifer Nelson explains how KP scientists are helping the CDC and FDA keep an eye out for rare adverse events.
mental health
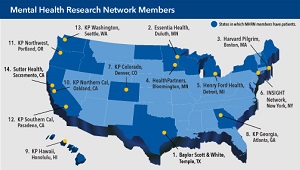
$10 million to expand Mental Health Research Network
NIMH funding will enable the MHRN to conduct larger studies in integrated health systems on topics that matter most.



