Health Informatics
Research overview
Research on health informatics at Kaiser Permanente Washington focuses on developing and using health information technology (IT) to transform health care delivery. By testing new paradigms of care that provide more opportunities to engage patients, this research is supplying valuable evidence that is helping shape federal policy and guiding innovative redesign of health care.
“We’re working to understand how to make health IT practical so patients and care teams find it useful and engaging,” explained Kaiser Permanente Washington Health Research Institute (KPWHRI) Senior Investigator James Ralston, MD, MPH. “We want to find ways to use information technologies to support patients and providers together, both inside and outside the office.”
Integral to this support is designing technologies that are user-friendly and meet the needs of both patients and providers. By applying human-centered methods that focus on needs, use, and usability, KPWHRI researchers inform the design of health IT with direct participation from users.
Groundbreaking methodological work by KPWHRI health informatics researchers includes developing natural language processing (NLP) to analyze text such as notes and written reports in electronic health records (EHRs). Assistant Investigator David Carrell, PhD, leads in the area of using NLP and machine learning to identify patient phenotypes, or specific health characteristics such as possible heart disease, risk of opioid overdose, or suggestion of colon cancer. This information can assist researchers in studying how genetics and other factors influence disease.
Other examples of KPWHRI health informatics research include projects using EHRs and secure electronic communications such as:
- using a patient-shared EHR to improve care for chronic illnesses such as depression, diabetes, hypertension, and heart disease;
- understanding the effects of technologies such as OpenNotes, which gives patients access to notes that their doctors write during office visits;
- understanding and addressing differences in patient use of online health care services that could lead to disparities in care;
- testing NLP to target mentions of specific words and phrases in EHRs to supplement or replace skilled chart abstraction—providing faster access to “big data” and actionable information about patients who may need follow up.
Examples of KPWHRI research in mobile health (mHealth) and user-centered design include:
- evaluating mHealth smartphone tools: 1) to improve primary care for alcohol use disorders, 2) to support patients after bariatric surgery, and 3) to change smoking behavior;
- the VITAL and Seeing Priorities studies to apply user-centered processes to learn how health care providers can elicit and honor what is most important to patients living with multiple chronic health conditions;
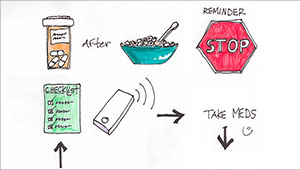
- the REMIND project applying user-centered methods to redesign clinical reminders and notifications for patients with chronic and preventive health care needs;
- the landmark Electronic Communications and Blood Pressure (eBP) study of home blood pressure monitoring and web-based care to increase hypertension control without office visits.
“Our studies on using health IT to improve care are showing that we can achieve better outcomes when we shift care from the doctor’s office to where people live: in their homes—and online,” said Senior Investigator Beverly B. Green, MD, MPH.
Recent publications on Health Informatics
Li R, Duan R, Zhang X, Lumley T, Pendergrass S, Bauer C, Hakonarson H, Carrell DS, Smoller JW, Wei WQ, Carroll R, Velez Edwards DR, Wiesner G, Sleiman P, Denny JC, Mosley JD, Ritchie MD, Chen Y, Moore JH Lossless integration of multiple electronic health records for identifying pleiotropy using summary statistics 2021 Jan 8;12(1):168. doi: 10.1038/s41467-020-20211-2. Epub 2021-01-08. PubMed
Jujjavarapu C, Anandasakaran J, Amendola LM, Haas C, Zampino E, Henrikson NB, Jarvik GP, Mooney SD ShareDNA: a smartphone app to facilitate family communication of genetic results 2021 Jan 6;14(1):10. doi: 10.1186/s12920-020-00864-0. Epub 2021-01-06. PubMed
Rosenthal EA, Crosslin DR, Gordon AS, Carrell DS, Stanaway IB, Larson EB, Grafton J, Wei WQ, Denny JC, Feng QP, Shah AS, Sturm AC, Ritchie MD, Pacheco JA, Hakonarson H, Rasmussen-Torvik LJ, Connolly JJ, Fan X, Safarova M, Kullo IJ, Jarvik GP Association between triglycerides, known risk SNVs and conserved rare variation in SLC25A40 in a multi-ancestry cohort 2021 Jan 6;14(1):11. doi: 10.1186/s12920-020-00854-2. Epub 2021-01-06. PubMed
Von Korff M, DeBar LL, Deyo RA, Mayhew M, Kerns RD, Goulet JL, Brandt C Identifying Multisite Chronic Pain with Electronic Health Records Data 2020 Dec 25;21(12):3387-3392. doi: 10.1093/pm/pnaa295. PubMed
Grant RW, McCloskey J, Hatfield M, Uratsu C, Ralston JD, Bayliss E, Kennedy CJ Use of Latent Class Analysis and k-Means Clustering to Identify Complex Patient Profiles 2020 Dec;3(12):e2029068. doi: 10.1001/jamanetworkopen.2020.29068. Epub 2020-12-01. PubMed
Researchers in Health Informatics
 Claire Allen, MPHManager, Collaborative Science |
 Katharine A. Bradley, MD, MPHSenior Investigator |
 Yates Coley, PhDAssociate Biostatistics Investigator |
 Beverly B. Green, MD, MPHSenior Investigator |
 Annie Hoopes, MD, MPHAssistant Investigator |
 Paula Lozano, MD, MPHSenior Investigator; Director, ACT Center |
 James D. Ralston, MD, MPHSenior Investigator |
 Brian D. Williamson, PhDAssociate Biostatistics Investigator |